BioHPC @ CBSU
BioHPC is the main CBSU venue
for delivering High Performance Computing to biological groups. It
was developed in collaboration with Microsoft and due to its
sponsorship part of CBSU resources are open to general scientific
community. BioHPC users are divided into two categories: guests and
registered users. Registered users come from Cornell community and
have several privileges over guests including number of jobs and
full access to restricted computationally expensive programs. BioHPC
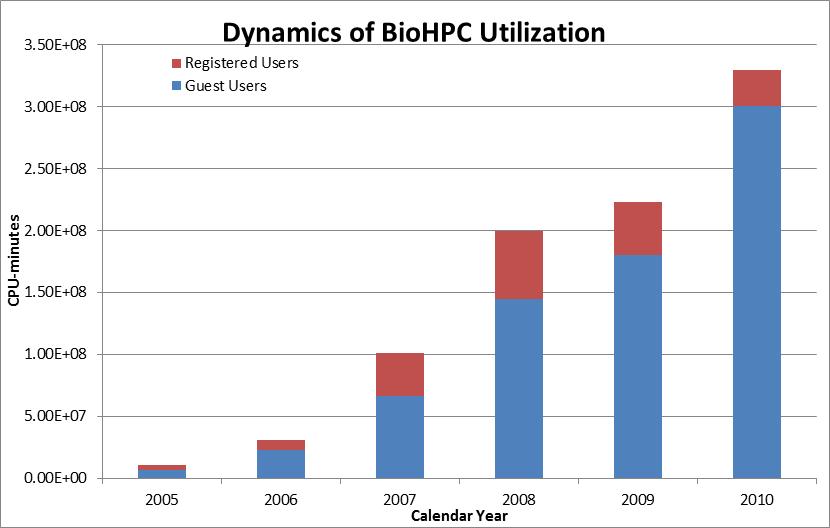
was very well received in the scientific community and its usage,
both registered and guest, increases substantially every year.
The Cornell BioHPC installation is available at
http://cbsuapps.tc.cornell.edu/ with all implemented
applications included. Some of the applications are freely available
for general public use; some, due to a very high computational
demand, are available to registered users only. Currently the CBSU
BioHPC installation at Cornell is linked to 327 compute nodes with
978 CPU cores grouped in 8 clusters. One compute node is a 64GB RAM
16-core server; 32 nodes are 8-core 16GB RAM servers; and, 234 nodes
are 2-core 4GB RAM servers. Twenty compute nodes from a 64 node
Athena cluster at Microsoft headquarters in Redmond, WA are linked
to this interface via a HPC Basic Profile/JSDL connection (16GB RAM
4-core servers). Due to size and load, the Cornell BioHPC
installation has a dedicated web server, separate fileserver (6TB
storage), separate ftp server (6TB storage) and a separate Microsoft
SQL Server. 2.5 FTEs at are currently dedicated to BioHPC
development and maintenance at CBSU.
The primary installation running on
Cornell's CBSU clusters processes over 80,000 job submissions per
year, the majority requiring multiple processors over multiple days.
Since its initial deployment in 2003 through 3/31/2011, BioHPC
processed 229,375 jobs with an average load of 27,597 jobs per year
and 86,151 over last full year (4/1/2010-3/31/2011). Jobs were
submitted by 16,224 users from 83 countries with the majority of
jobs (49% by CPU time used) originating in the U.S. Among them there
are 3,380 users from .edu domains representing 498 edu institutions,
7,230 users from .com domains (including 6,520 users with Yahoo,
Gmail and Hotmail e-mail addresses), and 311 Cornell users.
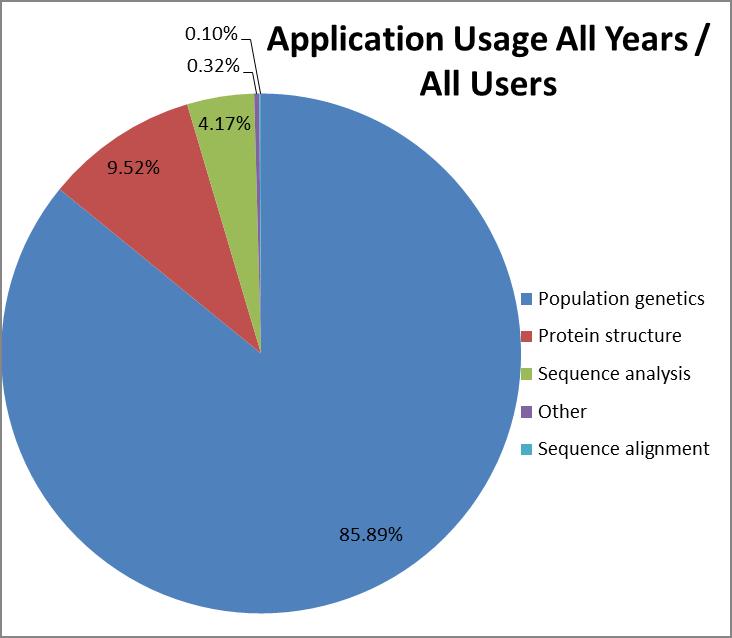
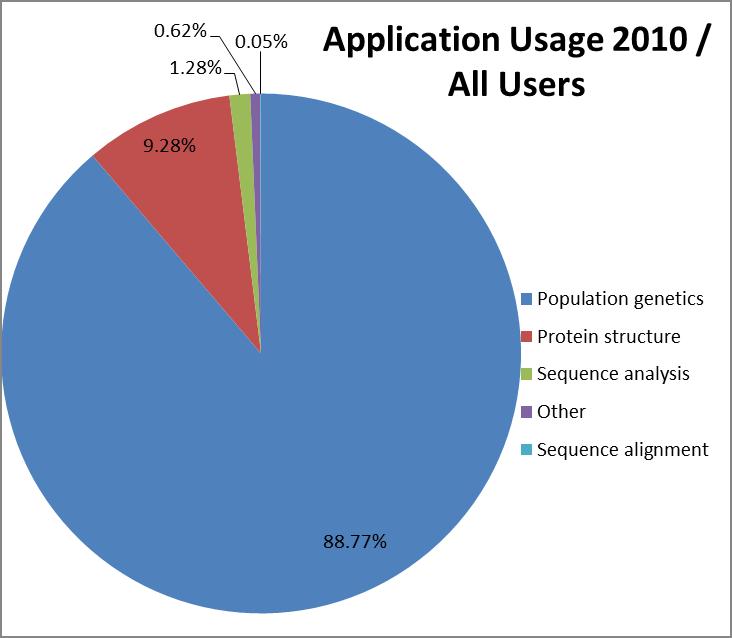
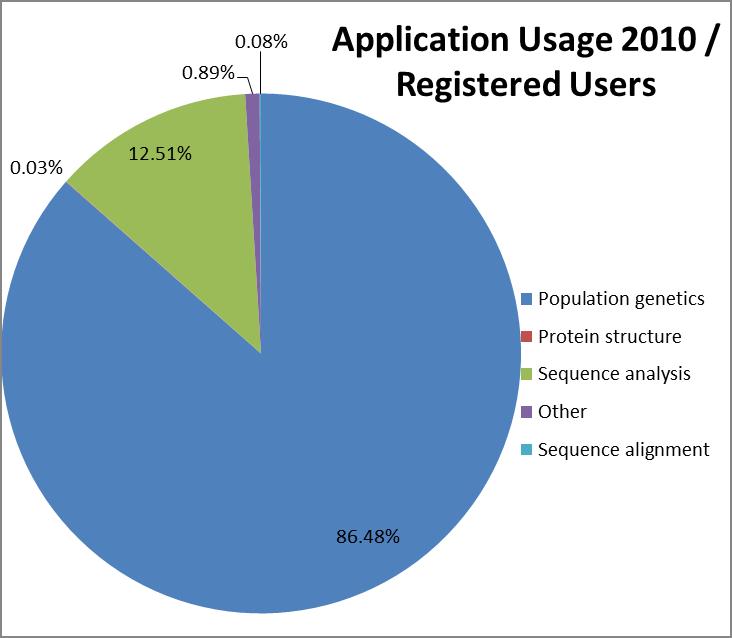
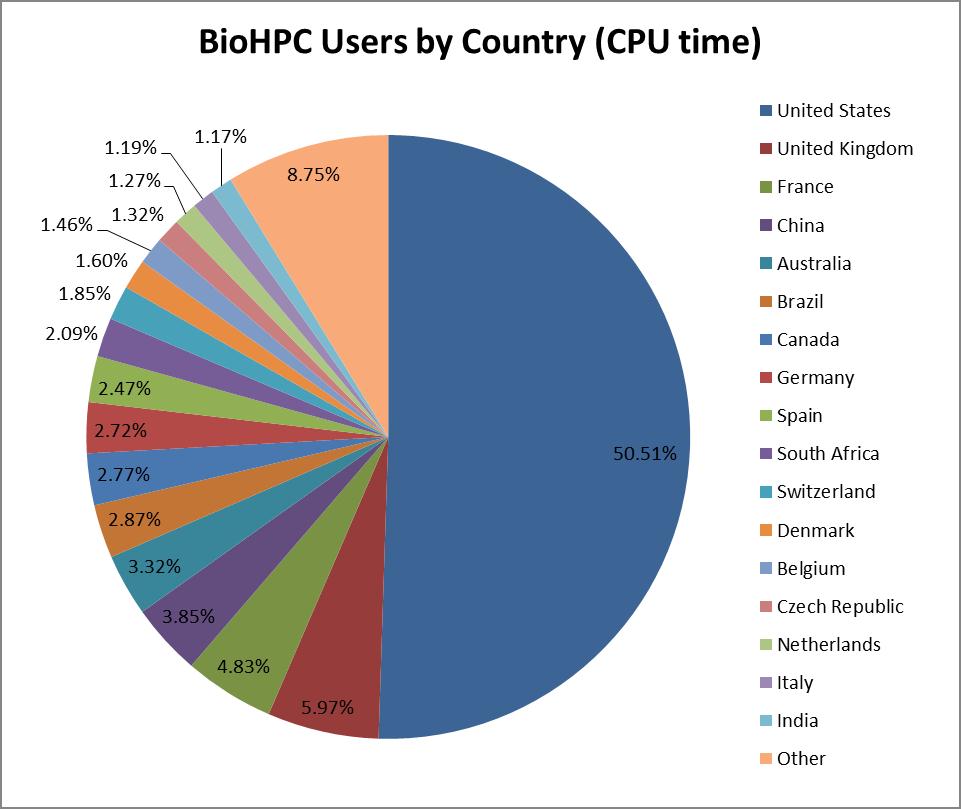
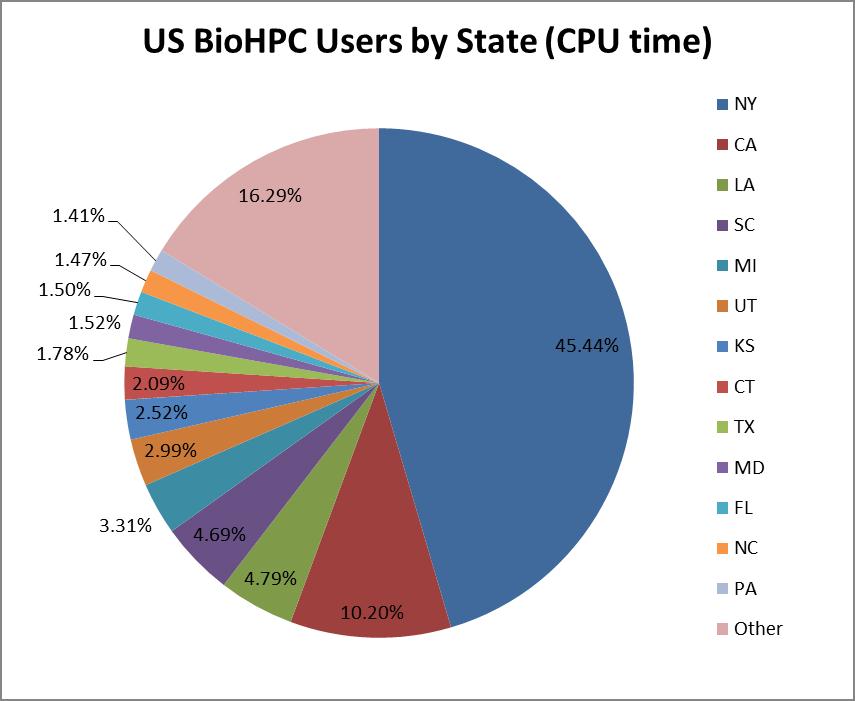
The most popular application category overall
(counting all years since 2003) is population genetics with 84% of
CPU time used followed by protein structure (10%) and sequence
analysis (5%).
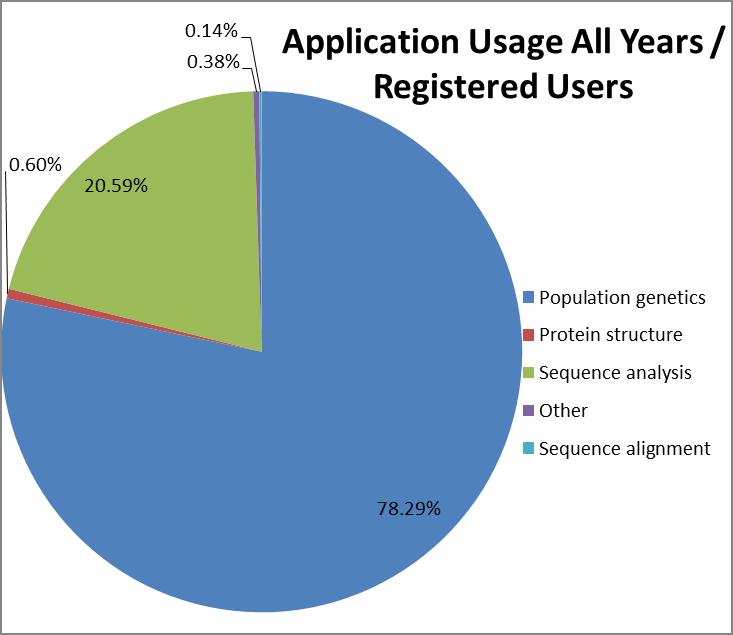
Cornell registered users
accounted for 24% of CPU usage while accounting for only 2.2% of the
total amount of users, which means Cornell users consume significant
computational resources per job and per user. Registered users have
access to the most resource consuming sequence analysis programs not
available to guests, however with majority of their usage still
focused on population genetics. They also can run more jobs per user
than guests resulting in higher resource utilization per category.
Recently, we gathered geographical
information of visitors of the Cornell BioHPC installation site
between June 3rd 2010 - May 19th 2011 using
RevolverMaps. It registered
89,306 visits with the following geographical distribution:
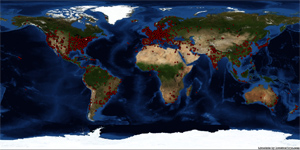
Visits are well correlated with job submissions.
Current visitors geographical distribution can be accessed
here.